Table of Contents
General Information
Gene Tools, LLC is the exclusive commercial source for research quantities of Morpholino oligos.
In Japan, Morpholino oligos are distributed soley by Funakoshi Co.
Morpholino oligos were devised by James Summerton in 1985 and were developed at ANTIVIRALS Inc. (now Sarepta Therapeutics Inc.), the pioneer antisense company founded by Summerton in 1980. James Summerton, Ph.D. is now the Manager of Gene Tools, LLC.
Morpholinos and the subunits used in their assembly have been awarded multiple pending and issued US and international patents, including particularly US Patents 5,142,047 and 5,185,444.
Design
By 1984 a growing concern in the then-emerging antisense field was that antisense therapeutics might never be commercially viable because of their very high production costs. Two key factors in the high cost of DNA analogs are: 1) the limited availability and high cost of their deoxyribonucleoside precursors; and, 2) the complexity and expense associated with coupling to hydroxyls, required in forming the phosphoester intersubunit linkages of most nucleic acid analogs.
In 1985 Summerton devised the Morpholino structural type to circumvent both of these cost problems [Summerton, 1989; Summerton, 1990]. This is achieved by starting with much less expensive ribonucleosides and introducing an amine via a relatively simple ribose-to-morpholine transformation. The resulting Morpholino subunits can be assembled into antisense oligos via simple and efficient coupling to the morpholine nitrogen, without the expensive catalysts and post-coupling oxidation steps required in the production of most nucleic acid analogs.
Early molecular modeling suggested that Morpholinos should effectively bind complementary nucleic acids. This predicted good binding affinity has been confirmed, as described later herein. Additionally, their unnatural structures and lack of anionic sites on the backbone were expected to preclude degradation by nucleases. This also has been confirmed by extensive experiments noted in a subsequent section.
Preparation
- Subunit Synthesis The first step in preparing Morpholino oligos is to convert ribonucleosides to Morpholino subunits as shown in Figure 1.
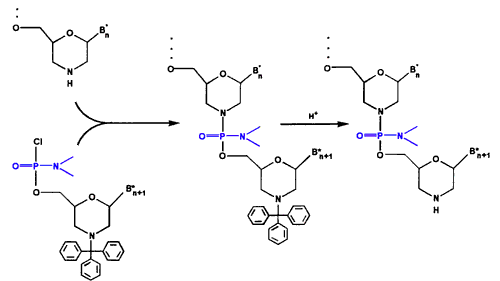
- Oligo Assembly As shown in Figure 3, we assemble the activated subunits into oligos of defined sequence on a solid-phase support using a simple two-reaction cycle, with intervening washes. The first reaction entails adding activated subunit. The second reaction entails detritylation to prepare the chain terminus for the next subunit addition.
- Oligo Processing Concentrated NH4OH is used to cleave the oligo from the resin and remove protective groups from the nucleobases. The finished oligo is cleaned up by filtration and selective precipitation. The oligo is resuspended in pure water. An aliquot is taken for MALDI-TOF mass spectrometry and another aliquot is quantitated spectrophotometrically at 265 nm in 0.1 N HCl. UV spectra are taken at acidic pH to preclude errors due to hypochromicity resulting from stacking of the bases.
Figure 1. Ribonucleoside to Morpholino Transformation
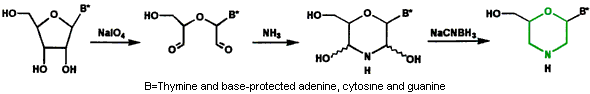
Morpholino subunits are subsequently protected with a trityl and activated with our phosphoroamidate linking agent, as shown in Figure 2.

Figure 2. Protection and Activation of Morpholino Subunit
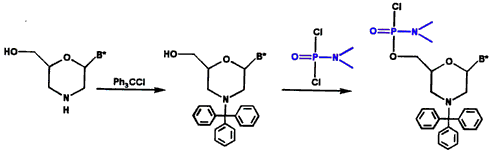

Figure 3. Oligo Assembly Cycle

The value from this UV spectral quantitation is used to calculate oligo concentration, after which 300 nanoMoles of oligo is placed into a vial and freeze dried. The vial is sealed and baked to give a sterile freeze-dried oligo.
To use that oligo, one simply adds a defined volume of sterile water to form a sterile pre-quantitated stock solution ready to use in antisense experiments.
Properties
- Solubility An antisense oligo should have good water solubility to assure good access to its target sequence within the cell. Conventional wisdom in the antisense field is that non-ionic antisense oligos (such as Morpholinos) invariably show poor water solubility. Contrary to that conventional wisdom, it has been found that if the nucleobases in a non-ionic antisense oligo stack poorly (determined by circular dichroism studies) then that oligo has poor water solubility, but if the nucleobases are well stacked in aqueous solution then that oligo can show excellent water solubility.
- Stability For optimal activity an antisense oligo should be completely resistant to nucleases. DNA antisense oligos are degraded in serum and within cells in a matter of minutes.
- Binding Affinity The thermal melt (Tm) results in Figure 4 illustrate the affinity of a Morpholino oligo (comprising A, C, G and U bases) for its complementary RNA sequence, as well as the affinities of corresponding S-DNA, DNA, and 2'O-Methyl RNA oligos for that same RNA sequence. As seen in this figure, the S-DNA has by far the lowest affinity, DNA has the next higher affinity, Morpholino a still higher affinity, and 2'O-Methyl RNA has the highest affinity. It should be noted that a corresponding PNA (Peptide Nucleic Acid)/RNA duplex would have a Tm quite similar to that of the 2'O-Methyl RNA/RNA duplex.
- Efficacy: Cell-Free It has been found that in a cell-free translation system with added RNase H (required for good S-DNA activity) the "old" 25-mer Morpholinos containing uracils typically exhibited slightly higher efficacies than corresponding S-DNAs, while "new" thymine-containing 25-mer Morpholinos generally achieve substantially higher efficacies than corresponding S-DNAs.
- Efficacy: In Cells In experiments with Morpholinos carried out at ANTIVIRALS Inc. it has been found that Morpholino antisense oligos which exhibit good activity in a cell-free translation system also exhibit correspondingly good activity when scrape-loaded into cultured animal cells [submitted for publication]. Similar correspondence between cell-free and in-cell activities of Morpholinos has been reported by Kobzik and coworkers at Harvard [Taylor et al., 1996] and by Kole and coworkers at Univ. of North Carolina [Ryszard Kole, personal communication], and by Giles at University of Liverpool.
This stacking/solubility correlation is evidenced by the poor stacking of carbamate-linked DNA analogs and carbamate-linked Morpholino oligos [Kang et al., 1992] and their corresponding poor aqueous solubilities [Stirchak, Summerton & Weller, 1987; Stirchak, Summerton & Weller, 1989]. In contrast, the standard non-ionic phosphorodiamidate-linked Morpholino oligos shown in Figure 1 exhibit excellent base stacking - even better than that in DNA [Kang et al., 1992] and they have very good water solubility - evidenced by disolution of 263 mg of a representative 22-mer in 1 ml of water without reaching saturation [Summerton & Weller, 1997a].
It is postulated that the low water solubility of non-ionic oligos with poorly-stacked bases is a consequence of the difficulty of inserting the hydrophobic faces of the unstacked bases into an aqueous environment. Conversely, the high water solubility of non-ionic phosphorodiamidate-linked Morpholinos is likely due to effective shielding of those hydrophobic faces from the polar solvent because of the exceptionally good stacking of the bases.
Currently-popular thiophosphates (S-DNAs) are appreciably more stable than DNA. Nonetheless, S-DNAs are substantially degraded over a period of hours to days - thereby seriously limiting their utility in antisense experiments running more than a few hours. Further, unpublished reports suggest that the thionucleotides resulting from degradation of S-DNAs are toxic to cells.
Chimeric oligos, which have recently become very popular, contain a segment of DNA or S-DNA (typically 8 to 12 bases) sufficient to afford RNase H cleavage of a paired target RNA. That RNase H-competent segment is flanked on one or both sides with an RNase H-independent segment designed to increase target binding affinity and/or increase resistance to exo nucleases. While chimeras have improved stability, it seems unlikely they will ever achieve satisfactory stability in biological systems because their RNase H-competent segment is inherently sensitive to endo nucleases within cells.
In sharp contrast to the limited stabilities inherent in the above RNase H-competent oligos (DNA, S-DNA, chimeras), Morpholino oligos have been shown to be completely resistant to nucleases, as well as being resistant to a broad range of other degradative factors in biological systems - even including a liver homogenate [Hudziak et al., 1996]. Accordingly, Morpholinos are effective in even long term experiments and they are free of complications which could arise from toxic degradation products.
Figure 4. Thermal Melt Profiles For Various Oligo/RNA Duplexes
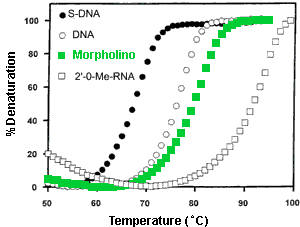
In 1998 Gene Tools, the sole commercial producer of research quantities of Morpholinos, switched from uracils to thymines in the production of Morpholinos. This increases the Tm a few degrees over that shown in Figure 5 and significantly increases efficacy. A "new" 25-mer containing thymines has comparable efficacy to a corresponding "old" 28-mer containing uracils. Switching from uracils to thymines also increases the Morpholino's ability to invade RNA secondary structure - thereby further improving its already outstanding targeting predictability.

In contrast to the case for low-affinity S-DNAs, Figure 5 shows that the 2'O-Methyl RNA/RNA duplex has an appreciably higher Tm than the corresponding Morpholino/RNA duplex. From this one might predict that 2'O-Methyl RNAs should have greater efficacies than corresponding Morpholinos. However, experiments suggest that in cell-free translations Morpholinos generally have slightly higher efficacies and substantially better specificities than corresponding 2'O-Methyl RNAs [ANTIVIRALS Inc. Technical Report 3, 1993].
The Tm values for PNA/RNA duplexes closely resemble the high Tm values for corresponding 2'O-Methyl RNA/RNA duplexes. However, contrary to the case for 2'O-Methyl RNAs, in cell-free studies with equal length PNAs and Morpholinos (18-mers) the higher-affinity PNAs generally achieve moderately higher efficacies than the corresponding lower-affinity Morpholinos. However, this efficacy advantage of PNAs is counterbalanced by the Morpholinos' substantially better sequence specificities.
In a more realistic analysis where the longest-commercially-available PNAs (18-mers) are compared to corresponding longest-commercially-available Morpholinos (25-mers), the longer Morpholinos achieve much higher efficacies than the shorter PNAs. Contrary to conventional wisdom which holds that specificity decreases with increasing length, the 25-mer Morpholinos also afford substantially higher sequence specificities than the 18-mer PNAs. Finally, the higher-efficacy/higher-specificity 25-mer Morpholinos are less expensive than the lower-efficacy/lower-specificity 18-mer PNAs.
A number of S-DNAs previously shown to have excellent efficacies in a cell-free test system were also tested for their efficacies within cells. In sharp contrast to the high in-cell efficacies of Morpholinos, the scrape-loaded S-DNAs typically showed only slight in-cell efficacy, and then only at concentrations hundreds of times higher than required for good activity by Morpholinos.
Initially it was suspected that lack of in-cell activity by scrape-loaded S-DNAs might be due to their multiple negative backbone charges preventing good cell entry during the scrape-load procedure. However, when fluorescein-labeled oligos were scrape-loaded into cells it was seen that this procedure achieves delivery of S-DNAs as well or better than delivery of Morpholinos [Summerton et al., 1997].
A possible explanation for the very poor in-cell efficacy we've seen with S-DNAs is that their complexes with target sequences are disrupted by some cellular factor. In this regard a factor in the nucleus has been reported which destabilizes RNA secondary structure [Pontius & Berg, 1992] and an RNA helicase has been reported which strand separates DNA/RNA duplexes [Flores-Rozas & Hurwitz, 1993]. Moulds et al. at Gilead Sciences have postulated that some such strand separating factor acts on antisense oligo/RNA duplexes within cells [Moulds et al., 1995] . This is based on experiments wherein they paired high-affinity anionic antisense oligos with their respective target RNAs and microinjected these duplexes into cells. They found that the oligos were efficiently stripped off their RNAs - evidenced by translation of the protein coded by that RNA.
It is postulated that this cellular strand-separating factor may strip off anionic S-DNAs but fail to act on corresponding oligo/RNA duplexes when the oligo is non-ionic - as is the case for Morpholinos. The premise is that most proteins which interact with nucleic acids do so in substantial part via ionic interactions with the anionic backbones of nucleic acids. Thus the strand-separating factor may simply lack an effective handle for interacting with the non-ionic Morpholino antisense oligos in oligo/RNA duplexes.
An additional contribution to the excellent in-cell efficacies of Morpholinos and the poor in-cell efficacies of S-DNAs in scrape-loaded cells may derive from their respective subcellular localization. Specifically, when low concentrations of fluorescein-labeled S-DNAs are scrape-loaded into cells fluorescence microscopy shows them to be largely sequestered in the nucleus. In contrast, fluorescein-labeled Morpholinos scrape-loaded into cells are distributed throughout the cytosol and nucleus.
To appreciate the impact these subcellular distributions may have on efficacy it should be noted that in the nucleus transcription of a pre-mRNA, processing of that pre-mRNA, and transport of the mature mRNA to the cytosol typically occurs within minutes. In contrast, translation of the mRNA in the cytosol typically continues for hours to days. As a consequence, S-DNAs sequestered in the nucleus may have access to their target RNAs only in the brief time between transcription and export to the cytosol, while the more homogeneously distributed Morpholinos should have access to their target RNAs during the RNAs' brief sojourn in the nucleus and during their far longer residence in the cytosol. It seems quite possible that this large difference in target access time might contribute substantially to the high in-cell efficacies of Morpholinos and the low in-cell efficacies of S-DNAs.
Specificity
A key factor which lured many scientists (and investors) into the antisense field was the expectation that an antisense therapeutic could, with near perfect specificity, block its targeted genetic sequence while exerting essentially no other effects on the patient [Summerton, 1979]. It was hoped such exquisite specificity by antisense therapeutics would avoid the severe toxicities characteristic of present small-molecule antiviral and anticancer therapeutics. These great expectations largely died for many scientists in the antisense field, particularly those in pharmaceutical research, when: a) it became widely (but erroneously) believed that RNase H-competency was essential for good efficacy; and, b) the severe specificity limitations of S-DNAs became widely appreciated.
While the challenges to developing antisense therapeutics which are both highly effective and highly specific are daunting, success in meeting these and other challenges may lead to safe and effective treatments for a broad range of currently intractable and devastating diseases.
To meet the specificity challenge antisense oligos should have a "minimum inhibitory length" (MIL) sufficiently long to avoid attack on essentially all nontarget sequences in the cellular RNA pool. The MIL is defined as the shortest length of oligo of a given structural type which achieves substantial target inhibition at concentrations typically achieved in the cytosol/nuclear compartment of treated cells. For antisense therapeutics in humans the RNA pool includes all of the RNA species in all of the different tissues of the patient. This is estimated to be about 200 million bases of unique-sequence RNA, based on about 6% to 7% of the genome being transcribed in humans post embryogenesis.
The challenge for RNase H-competent oligos (ie., DNA, S-DNA, and chimerics) is to distinguish a single selected target sequence from this vast pool of RNA sequences.
Happily, the challenge for RNase H-independent oligos, such as Morpholinos, is less daunting. This is because RNase H-independent types are inactive when targeted against the approximately 95% to 98% of cellular RNA sequences comprising intron sequences in pre-mRNAs and amino acid-coding and 3'untranslated sequences more than about 20 bases 3' to the translational start site in mRNAs. As a consequence, Morpholinos should enjoy an inherent 20-fold to 50-fold specificity advantage over S-DNAs and chimerics because Morpholinos are only required to distinguish their target sequences from about 2% to 5% of the sequences comprising the patient's RNA pool.
Table 1 illustrates the importance of an adequate MIL value for achieving good sequence specificity. This table tabulates MIL values and the corresponding number of inherent cellular RNA sequences expected to be inadvertently inactivated by 25-mer oligos of the two fundamental antisense classes: RNase H-competent (eg., S-DNAs and chimerics) and RNase H-independent (eg., Morpholinos and PNAs). In calculating the values in this table an estimated RNA pool size of 200 million unique-sequence bases was used, as well as an estimate that sequences targetable by RNase H-independent oligos comprise 5% of that RNA pool.
| MIL | RNase H Competent |
RNase H Independent |
|---|---|---|
| 7 | 232,000 | 11,600 |
| 8 | 55,900 | 2,750 |
| 9 | 13,000 | 648 |
| 10 | 3,050 | 153 |
| 11 | 715 | 36 |
| 12 | 167 | 8 |
| 13 | 38 | 2 |
| 14 | 9 | 0 |
| 15 | 2 | 0 |
| 16 | 0 | 0 |
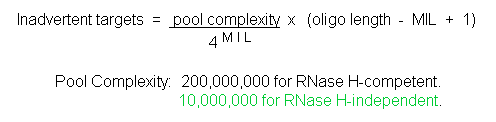
Experiments have been carried out at ANTIVIRALS Inc. (now Sarepta Therapeutics) to estimate MIL values for both RNase H-competent S-DNAs and RNase H-independent Morpholinos. In these activity versus length experiments both S-DNA and Morpholino oligos ranging in length from 8 bases to 30 bases, all targeted against the same region of rabbit alpha-globin leader sequence, were assessed in a cell-free translation system for their abilities to inhibit translation of a downstream luciferase coding sequence [for specifics on this test system see: [Summerton et al., 1997]. Figure 5 shows the respective translational inhibition values at oligo concentrations of 300 nanoMolar.
Figure 5. Activity Versus Oligo Length In Cell-free Translation System With 300 nM Oligo
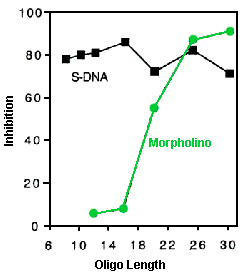
The results in Figure 5 suggest that under the conditions of this experiment S-DNAs have an MIL value of less than 8. This is far short of the minimum of 15 to 16 estimated to be required for high sequence specificity by RNase H-competent oligos. In sharp contrast, Morpholinos in this experiment have an MIL value of about 16. This is comfortably over the minimum of 13 to 14 expected to be required for high sequence specificity by RNase H-independent oligos.

The values in Table 1 combined with the results in Figure 5 suggest that every S-DNA may have a large number of inadvertent targets in human cells. In contrast, any given Morpholino is expected to have essentially no inadvertent targets.
To test these predictions, highly stringent specificity assays have been carried out in a cell-free translation system. In these experiments two oligos of each structural type were used. One oligo of each type was perfectly complementary to its target mRNA to provide a measure of total inhibition achieved by that oligo type. The other oligo of that type incorporated 4 mispairs to that same target sequence, with the longest run of perfect pairing comprising 10 contiguous base pairs, to provide an emulation of the estimated level of sequence homology likely to be encountered in the RNA pool within human cells. The difference between these two inhibition values at each concentration, defined as "high-specificity inhibition", provides a concentration-dependent measure of the high-specificity inhibition achieved by that structural type. Results from a representative experiment are shown in Figure 6 [experimental as in [Summerton et al., 1997].
Figure 6. High-Specificity Inhibition In Cell-free Translation System
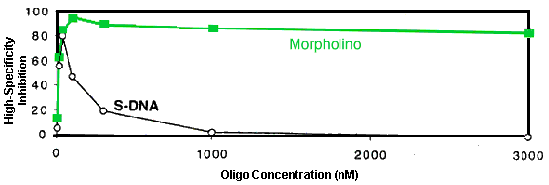
As predicted from the MIL values in Table 1 and the length versus activity results in Figure 12, the results in Figure 6 demonstrate that the RNase H-competent S-DNA achieved little high-specificity inhibition (47% to 80%) and then only in a quite narrow concentration range (10 nanoMolar to 100 nanoMolar). In contrast, but also in accord with these predictions, the RNase H-independent Morpholino achieved excellent high-specificity inhibition (84% to 95%) over a vastly broader concentration range (30 nanoMolar to well over 3,000 nanoMolar).

These results suggest that the long sought "exquisite specificity" goal of antisense has been achieved by Morpholinos. In contrast, because of the basic mechanism underlying RNase H-competency it is unlikely RNase H-competent oligos (S-DNAs and chimerics) can ever achieve a commensurate level of specificity.
Commercial Source
Gene Tools, LLC markets research quantities of Morpholino oligos worldwide. Our website has information on Morpholino products, prices, and ordering.
You can contact us electronically or by using the information at the bottom of every page on this site.
In Japan, Funakoshi Co. is the distributor of Morpholino oligos and related products.
References
- ANTIVIRALS Technical Report 3 (1993). Comparative studies of efficacy and specificity of translation inhibition in reticulocyte lysates. Antisense Res. Dev. 3 , following p. 306.
- Flores-Rozas & Hurwitz (1993). Characterization of a new RNA helicase from nuclear extracts of HeLa cells which translocates in the 5' to 3' direction. J. Biol. Chem. 268 , 21372.
- Hudziak, Barofsky, Barofsky, Weller, Huang, Weller (1996). Resistance of Morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 6 , 267.
- Kang, Cho, Johnson, Weller, Huang, Summerton (1992). Stacking interactions of ApA analogues with modified backbones. Biopolymers 32 , 1351.
- Moulds, Lewis, Froehler, Grant, Huang, Milligan, Matteucci, Wagner (1995). Site and mechanism of antisense inhibition by C-5 propyne oligonucleotides, Biochem. 34 , 5044.
- Pontius & Berg (1992). Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J. Biol. Chem. 267 , 13815.
- Stirchak, Summerton & Weller (1987). Uncharged stereoregular nucleic acid analogues. 1. Synthesis of a cytosine-containing oligomer with carbamate intersubunit linkages. J. Organic Chemistry 52 , 4202.
- Summerton (1989). Uncharged nucleic acid analogs for therapeutic and diagnostic applications: oligomers assembled from ribose-derived subunits. In: Discoveries in Antisense Nucleic Acids; Ed.: C. Brakel; Pub.: The Portfolio Publishing Co., Woodlands, Texas; pages 71 - 80.
- Summerton & Weller (1997a). Morpholino antisense oligomers: design, preparation and properties. Antisense Nuc. Acid Drug Dev. 7 , 187.
- Taylor, Paulauskis, Weller, Kobzik (1996). In vitro efficacy of Morpholino-modified antisense oligomers directed against tumor necrosis factor-alpha mRNA. J. Biol. Chem. 271, 17,445.
