Gene Tools customer support personnel have tools and techniques to select Morpholino oligo sequences likely to produce good knockdowns or splice modifications. Morpholino oligo targeting is a free service available from Gene Tools. After target mRNA sequences are submitted for design, proposed Morpholino antisense sequences are usually returned by email during the next business day.
Answering a few questions about your experimental system will help you design a Morpholino sequence both specific and highly effective against the target of interest. You do not need thorough answers for all of these questions to design great Morpholino oligos, however, your chance of success increases the more you know about your target. Relevant questions include: What is the structure of the mRNA or pre-mRNA? Are there spliceoforms or splice-sites I would like to block? Do I have enough sequence around the start codon and upstream to design one or more translational-blocking Morpholino oligos? Does the target sequence share homology with undesirable gene targets? What are the experimental conditions (e.g. temperature and time constraints) for my study? In addition to the targeting questions above, you should keep in mind these two questions important for overall experimental design: How will I assay for successful gene knockdown? How will I deliver the oligos into the cytosol of cells?
Once you've found a viable target region and chosen an effective cytosolic delivery method and a method to assay for antisense activity, it is the Morpholino oligo sequence that will determine success. Importantly, the oligo must have a base sequence that has very little self-complementarity so that it does not dimerize. Otherwise, the optimal Morpholino oligo will be about 25-bases long and have a high enough GC content (e.g. 40-60%) so that it has a high target affinity. The oligo should not have stretches of four or more contiguous G so that remains water soluble.
In the most typical scenario the complete mRNA sequence is known and one wishes to block translation. For this application, an optimal target is a 25-base sequence that lies within the region from the 5' cap through the first 25-bases of coding sequence, has a ~50% GC content and has little-or-no secondary structure. Except in the unusual circumstance of a gene with an internal ribosome entry site (IRES), there is no particular advantage to targeting the start codon instead of upstream sequence.
Usually one can find at least 2 or 3 oligo targets within the 5' cap to start region that have roughly equivalent properties. Oligos complementary to any of these targets should reduce expression of the downstream coding region. In any set of oligos targeted to a given gene there will likely be a "best" oligo that achieves the greatest knockdown, sometimes to an undetectable level of gene expression. In many cases trying just one oligo is enough for effective knockdown. No other antisense structural type achieves the high rate of successful first-try knockdown enjoyed by users of Morpholino oligos.
The high success rate for Morpholino oligo targeting is likely due to their higher affinity for RNA. Morpholinos invade RNA secondary structures far more successfully than the low-affinity S-DNAs. A chronic problem in the antisense field has been the difficulty in predicting effective targets for antisense oligos. Specifically, S-DNAs are inactive against many potential target sequences in a selected RNA. This is likely because the weak-binding S-DNAs cannot invade relatively stable secondary structures in the RNA. Even when suitable single-stranded regions in the RNA are available for pairing, the low-affinity S-DNAs are often incapable of invading adjacent double-stranded regions. Routinely, sets of S-DNA oligos are screened to find a single oligo capable of knocking down its target gene. Routinely, Morpholino users buy one oligo and knock down one gene; about 80% of the time, the first Morpholino tried does the job.
The good targeting predictability of Morpholinos is demonstrated by experiments illustrated in Figures 1a and 1b. The mRNA targeted in these experiments has quite stable secondary structure but Morpholino oligos still achieved good knockdown. Figure 1a shows an mRNA construct comprising 85 bases of the leader sequence of Hepatitis B virus (HBV) mRNA joined to the amino acid coding sequence of firefly luciferase. The numbered lines indicate target sites for 7 different Morpholino antisense oligos. Figure 1b shows the translation knockdown achieved in a cell-free translation assay with 1 nanoMolar target mRNA. Each Morpholino oligo shown in Figure 1a was used at 1 microMolar concentration and their knockdown efficacy is plotted in Figure 1b as a function of their position along the mRNA, along with corresponding values for another 5 oligos positioned further 3' to the translational start site. It is particularly noteworthy that oligos 3, 4 and 5 are seen to have effectively invaded the quite stable secondary structure within the HBV leader sequence.
The results in Figure 1b suggest that by following rational and reliable targeting guidelines, Morpholino antisense oligos can be selected which have a high probability of being effective. These targeting guidelines are detailed below.
Blocking Translation: The following guidelines generally lead to Morpholino oligos which are highly effective for blocking translation of their targeted mRNAs.
1. Select a target sequence in the post-spliced mRNA in the region from the 5'cap to about 25 bases 3' to the AUG translational start site. As shown in Figure 2, Morpholinos (acting via a steric block mechanism) targeted more than about 30 bases 3' to the AUG translational start site do not block translation.
Figure 4 shows oligo A, having a barely acceptable 16 contiguous base-pairs of self- complementarity (2+3+3+3+3+2=16 contiguous hydrogen bonds), and oligo B, which should not be selected because it contains an unacceptable 18 contiguous base-pairs (3+3+3+3+3+3=18 contiguous hydrogen bonds) of self-complementarity.
3. A 25-base Morpholino oligo will likely have poor water solubility if it contains more than 9 total guanines (>36% G) or more than 3 contiguous guanines.
4. 25-Mers (the longest commercially available) are recommended for most applications. This is because efficacies increase substantially with increasing length and because long oligos best assure access to a single-stranded region in the target RNA, as is required for nucleation of pairing by the oligo.
Figure 5 demonstrates in a cell-free translation system the great gains in efficacy with increasing length of Morpholino oligos.
Blocking Nuclear Processing: Morpholino oligos can block nuclear processing events, in particular pre-mRNA splicing; the following guidelines have proven effective for designing oligos targeting splice sites. Blocking nuclear processing with Morpholino oligos provides some significant advantages over translational blocking. First, you can often create specific desired effects such as exon deletion or intron insertion by following some general oligo design guidelines. Second, all outcomes can be characterized and quantitated by RT-PCR; characterization of translation blocking by Western blotting must await protein degradation before clear knockdown can be measured, but RT-PCR of splice-blocked mRNA allows rapid assay for the new shifted band and, due to the short half-life of RNA compared with protein, the mRNA spliced prior to Morpholino treatment disappears rapidly.
In terms of composition and sequence motifs, an optimal splice-blocking oligo has the same properties described above for a translation-blocking oligo. However, splice-blocking oligos have additional target-specific requirements, the most important of which is a defined pre-mRNA sequence with explicitly known exon-intron and intron-exon boundaries. Figure 6 depicts a "typical" multi-exon pre-mRNA target.
In order to produce an exon deletion, a minimum of 3 exons is required in the pre-mRNA so that there is an internal exon as well as a first and last exon. Targeting exon-intron boundaries or intron-exon boundaries of internal exons generally results in complete or partial deletion of the abutting exon along with its flanking introns, as shown in the upper part of Figure 7. However, in cases where there is a great deal of divergence in splice sites from consensus, it is also possible to generate intron insertions. It is thought that these insertions occur because the unblocked splice boundaries at the far ends of the introns flanking a splice blocked exon are unable to be brought together, possibly due to poor binding of small nuclear ribonuclear proteins (snRNPs).
Targeting a splice junction of the first or last exon in a pre-mRNA generally triggers a complete or partial intron inclusion. We think that the reason these insertions occur is that the snRNP bound across the intron from the blocked exon junction is left partnerless when the Morpholino oligo prevents the snRNP's normal splice partner from binding and because there is no splice junction on the far side of a first or last exon, the remaining snRNP does not have another splice junction to which it can splice.
In Figure 6, the e1i1 or i3e4 Morpholinos would likely cause insertion of introns 1 or 3 respectively, the i1e2 or e2i2 Morpholinos would likely cause deletion of exon 2, and the i2e3 or e3i3 Morpholinos would likely cause deletion of exon 3; however as discussed above, some oligos expected to trigger deletions can trigger insertions instead and as discussed below, activation of cryptic splice sites could turn insertions into partial insertions or deletions into partial deletions.
Figure 8 shows a range of possible outcomes of blocking an internal exon, including exon skipping (the most common outcome), intron insertion, and the products of cryptic splice site activation, partial exon deletion and partial intron insertion. The figure shows a Morpholino targeting the donor site; similar outcomes are possible by blocking an acceptor site, though cryptic site activation is less likely because the sequence of a functional acceptor site is more constrained.
In summary, a Morpholino oligo directed against any splice junction is expected to generate either a complete or partial single exon deletion or a complete or partial single intron insertion. To use RT-PCR to analyze the mRNA product generated by a splice-blocking Morpholino, make a single set of primers targeted to sequence outside (5' and 3') of the predicted deletion or insertion. The Gene Tools customer support staff will be glad to help you both design your oligos and help you determine the best approach for your experiments. Feel free to call or email us.
Limitations: The most significant limitations for using Morpholino oligos fall into two categories, design and delivery. However, the constraints on design and challenge of delivery are only limitations for the unaware.
Regarding design, as long as appropriate regions of the mRNA or pre-mRNA are targeted and problematic moieties such as self-complementarity and runs of G are avoided the oligos are very likely to be effective. But it is important to remember that because Morpholino oligos work by a steric-blocking mechanism, effective oligos must either target sequence to block translation, pre-mRNA modification (e.g. splice jinctions), or regulatory protein binding (e.g. intronic splice suppressors, exonic splice enhancers).
As far as delivery is concerned, the non-charged nature of the Morpholino backbone has historically contributed to difficulty in delivery because Morpholinos cannot be delivered with cationic lipid reagents (lipofection reagents). For cells in culture, delivery of Morpholinos using our Endo-Porter reagent offers good delivery efficacy with less toxicity than when lipofection reagents are used to deliver nucleic acids. Electroporation is also a good method for introducing Morpholinos into cells. Of course microinjection of Morpholinos into embryos has proven to be very effective. It should be noted that delivery has been achieved in nearly all of the most widely studied biological systems. We suggest you look at our database of publications which allows you to browse all organisms studied.
As long as you know the characteristics of good design and the techniques for delivery and plan these into your experiments, we are confident you will achieve the cleanest knockdown available with unsurpassed specificity, minimal non-antisense effects, and amazingly low toxicity.
Introduction
Targeting Morpholino oligos is vastly simpler than targeting DNA-based antisense oligos. Morpholinos usually work on the first attempt if they are successfully delivered to the cytosol. DNA and S-DNA oligos have a low probability of shutting down a target gene on the first attempt; it is common for five or six oligos to be tried before an effective sequence is found. This is in part because a Morpholino oligo has a higher binding affinity than an equivalent DNA-based antisense oligo. This higher affinity allows Morpholinos to invade RNA secondary structure and substantially increases the probability of designing effective oligos. Precisely targeting a single gene is also simplified because Morpholino oligos act via steric-blocking as opposed to an RNAse H-mediated mRNA degradation mechanism. We will describe the effects of affinity and steric-blocking further in this section.
Answering a few questions about your experimental system will help you design a Morpholino sequence both specific and highly effective against the target of interest. You do not need thorough answers for all of these questions to design great Morpholino oligos, however, your chance of success increases the more you know about your target. Relevant questions include: What is the structure of the mRNA or pre-mRNA? Are there spliceoforms or splice-sites I would like to block? Do I have enough sequence around the start codon and upstream to design one or more translational-blocking Morpholino oligos? Does the target sequence share homology with undesirable gene targets? What are the experimental conditions (e.g. temperature and time constraints) for my study? In addition to the targeting questions above, you should keep in mind these two questions important for overall experimental design: How will I assay for successful gene knockdown? How will I deliver the oligos into the cytosol of cells?
Once you've found a viable target region and chosen an effective cytosolic delivery method and a method to assay for antisense activity, it is the Morpholino oligo sequence that will determine success. Importantly, the oligo must have a base sequence that has very little self-complementarity so that it does not dimerize. Otherwise, the optimal Morpholino oligo will be about 25-bases long and have a high enough GC content (e.g. 40-60%) so that it has a high target affinity. The oligo should not have stretches of four or more contiguous G so that remains water soluble.
In the most typical scenario the complete mRNA sequence is known and one wishes to block translation. For this application, an optimal target is a 25-base sequence that lies within the region from the 5' cap through the first 25-bases of coding sequence, has a ~50% GC content and has little-or-no secondary structure. Except in the unusual circumstance of a gene with an internal ribosome entry site (IRES), there is no particular advantage to targeting the start codon instead of upstream sequence.
Usually one can find at least 2 or 3 oligo targets within the 5' cap to start region that have roughly equivalent properties. Oligos complementary to any of these targets should reduce expression of the downstream coding region. In any set of oligos targeted to a given gene there will likely be a "best" oligo that achieves the greatest knockdown, sometimes to an undetectable level of gene expression. In many cases trying just one oligo is enough for effective knockdown. No other antisense structural type achieves the high rate of successful first-try knockdown enjoyed by users of Morpholino oligos.
The high success rate for Morpholino oligo targeting is likely due to their higher affinity for RNA. Morpholinos invade RNA secondary structures far more successfully than the low-affinity S-DNAs. A chronic problem in the antisense field has been the difficulty in predicting effective targets for antisense oligos. Specifically, S-DNAs are inactive against many potential target sequences in a selected RNA. This is likely because the weak-binding S-DNAs cannot invade relatively stable secondary structures in the RNA. Even when suitable single-stranded regions in the RNA are available for pairing, the low-affinity S-DNAs are often incapable of invading adjacent double-stranded regions. Routinely, sets of S-DNA oligos are screened to find a single oligo capable of knocking down its target gene. Routinely, Morpholino users buy one oligo and knock down one gene; about 80% of the time, the first Morpholino tried does the job.
The good targeting predictability of Morpholinos is demonstrated by experiments illustrated in Figures 1a and 1b. The mRNA targeted in these experiments has quite stable secondary structure but Morpholino oligos still achieved good knockdown. Figure 1a shows an mRNA construct comprising 85 bases of the leader sequence of Hepatitis B virus (HBV) mRNA joined to the amino acid coding sequence of firefly luciferase. The numbered lines indicate target sites for 7 different Morpholino antisense oligos. Figure 1b shows the translation knockdown achieved in a cell-free translation assay with 1 nanoMolar target mRNA. Each Morpholino oligo shown in Figure 1a was used at 1 microMolar concentration and their knockdown efficacy is plotted in Figure 1b as a function of their position along the mRNA, along with corresponding values for another 5 oligos positioned further 3' to the translational start site. It is particularly noteworthy that oligos 3, 4 and 5 are seen to have effectively invaded the quite stable secondary structure within the HBV leader sequence.
Figure 1. RNA Structure And Morpholino Antisense Activity
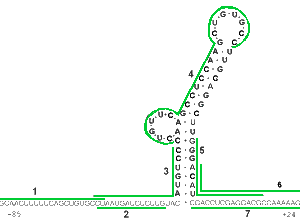
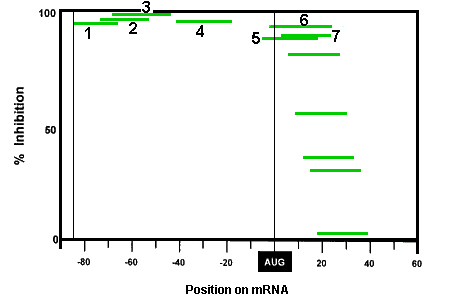
It should be noted that the low efficacy of Morpholinos targeted more than a few bases 3' to the AUG translational start site is probably a consequence of the unwindase activity associated with ribosome complex after full assembly at the translational start site. As described in our section on specificity, this lack of efficacy against sequences more than a few bases 3' to the translational start site actually contributes substantially to the greater specificity of Morpholinos relative to S-DNAs, RNAi and chimeric oligos.


The results in Figure 1b suggest that by following rational and reliable targeting guidelines, Morpholino antisense oligos can be selected which have a high probability of being effective. These targeting guidelines are detailed below.
Targeting Guidlines
Now we will take you through the steps of designing translation-blocking Morpholino oligos. Then we will show you how to design Morpholino oligos to block or alter mRNA splicing events. These targeting guidelines are the same as used by Gene Tools customer support staff when researchers request our free oligo design service.
Blocking Translation: The following guidelines generally lead to Morpholino oligos which are highly effective for blocking translation of their targeted mRNAs.
1. Select a target sequence in the post-spliced mRNA in the region from the 5'cap to about 25 bases 3' to the AUG translational start site. As shown in Figure 2, Morpholinos (acting via a steric block mechanism) targeted more than about 30 bases 3' to the AUG translational start site do not block translation.
Figure 2. Antisense Activity Versus Target Position In mRNA

A 25-base region that includes the translational start site is a simple and generally effective target, as long as there is not prohibitive self-complementarity or other undesirable motifs. When Gene Tools designs a Morpholino oligo for translation blocking, we start by analyzing the first 25 bases of coding sequence (including the start codon) and then slide the 25-base window upstream until a 25-base target is found that satisfies the general requirements for an optimal Morpholino oligo (i.e. 40-60% GC content without significant self-complementarity or stretches of 4 or more contiguous G). An advantage of approaching oligo targeting in this manner is that additional viable targets likely remain upstream in the 5' UTR. This becomes important when the researcher wishes to verify their antisense results with a second or third non-overlapping Morpholino oligo against the same target mRNA. We find that many researchers in developmental biology have preferred this "2 to 3 oligo strategy" to verify their antisense results. Figure 3 reiterates the limits of the acceptable region in which to select a target sequence for translational blocking.

Figure 3. Initial choice for a Targetable Region for translational blocking in mRNA
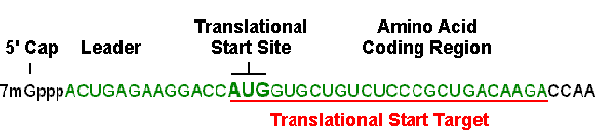
2. Make sure the selected sequence has little or no self-complementarity. The Morpholino sequence should be chosen so that the stability of self-complementary sequences is minimized; the selected Morpholino oligo should form no more than 16 contiguous intrastrand hydrogen bonds and preferably less. In some cases 16 contiguous base-pairs can be detrimental to achieving good antisense results if the resulting pairing is entirely between G and C residues.
Figure 4 shows oligo A, having a barely acceptable 16 contiguous base-pairs of self- complementarity (2+3+3+3+3+2=16 contiguous hydrogen bonds), and oligo B, which should not be selected because it contains an unacceptable 18 contiguous base-pairs (3+3+3+3+3+3=18 contiguous hydrogen bonds) of self-complementarity.
Figure 4. Oligos With Differing Self-complementarity
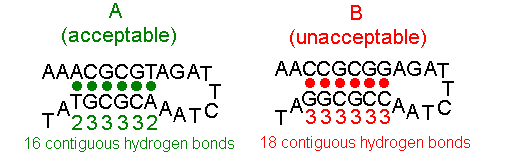
Note that the figures shows an intrastrand pairing event and that oligo B will form Morpholino dimers through interstrand pairing as well.

3. A 25-base Morpholino oligo will likely have poor water solubility if it contains more than 9 total guanines (>36% G) or more than 3 contiguous guanines.
4. 25-Mers (the longest commercially available) are recommended for most applications. This is because efficacies increase substantially with increasing length and because long oligos best assure access to a single-stranded region in the target RNA, as is required for nucleation of pairing by the oligo.
Figure 5 demonstrates in a cell-free translation system the great gains in efficacy with increasing length of Morpholino oligos.
Figure 5. Length Versus Activity For Morpholino Oligos
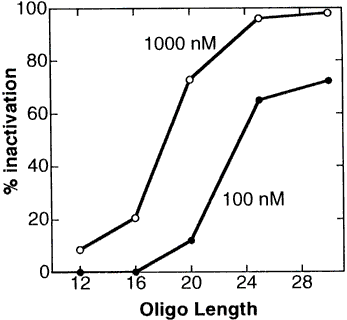
This length versus activity study was carried out with Morpholino oligos containing uracils. In 1998 Gene Tools switched from uracils to thymines, which increased the target binding affinity such that a "new" thymine-containing 25-mer has about the same efficacy as an "old" uracil-containing 28-mer. Morpholino oligos continue to be synthesized with thymines. From this study it can be seen that short Morpholinos (shorter than about 20 subunits) exhibit only modest efficacy, while longer Morpholinos (available in lengths up to 25-mer) are far more effective. Contrary to conventional wisdom in the antisense field, increasing the length of Morpholinos causes no significant loss in their specificity. This is described in detail in our section on Morpholino Oligo specificity. In contrast to Morpholinos or other steric-blocking oligos, the S-DNA oligos and other RNAse-H-dependant oligos do significantly decrease specificity with increasing length.

Blocking Nuclear Processing: Morpholino oligos can block nuclear processing events, in particular pre-mRNA splicing; the following guidelines have proven effective for designing oligos targeting splice sites. Blocking nuclear processing with Morpholino oligos provides some significant advantages over translational blocking. First, you can often create specific desired effects such as exon deletion or intron insertion by following some general oligo design guidelines. Second, all outcomes can be characterized and quantitated by RT-PCR; characterization of translation blocking by Western blotting must await protein degradation before clear knockdown can be measured, but RT-PCR of splice-blocked mRNA allows rapid assay for the new shifted band and, due to the short half-life of RNA compared with protein, the mRNA spliced prior to Morpholino treatment disappears rapidly.
In terms of composition and sequence motifs, an optimal splice-blocking oligo has the same properties described above for a translation-blocking oligo. However, splice-blocking oligos have additional target-specific requirements, the most important of which is a defined pre-mRNA sequence with explicitly known exon-intron and intron-exon boundaries. Figure 6 depicts a "typical" multi-exon pre-mRNA target.
In order to produce an exon deletion, a minimum of 3 exons is required in the pre-mRNA so that there is an internal exon as well as a first and last exon. Targeting exon-intron boundaries or intron-exon boundaries of internal exons generally results in complete or partial deletion of the abutting exon along with its flanking introns, as shown in the upper part of Figure 7. However, in cases where there is a great deal of divergence in splice sites from consensus, it is also possible to generate intron insertions. It is thought that these insertions occur because the unblocked splice boundaries at the far ends of the introns flanking a splice blocked exon are unable to be brought together, possibly due to poor binding of small nuclear ribonuclear proteins (snRNPs).
Targeting a splice junction of the first or last exon in a pre-mRNA generally triggers a complete or partial intron inclusion. We think that the reason these insertions occur is that the snRNP bound across the intron from the blocked exon junction is left partnerless when the Morpholino oligo prevents the snRNP's normal splice partner from binding and because there is no splice junction on the far side of a first or last exon, the remaining snRNP does not have another splice junction to which it can splice.
In Figure 6, the e1i1 or i3e4 Morpholinos would likely cause insertion of introns 1 or 3 respectively, the i1e2 or e2i2 Morpholinos would likely cause deletion of exon 2, and the i2e3 or e3i3 Morpholinos would likely cause deletion of exon 3; however as discussed above, some oligos expected to trigger deletions can trigger insertions instead and as discussed below, activation of cryptic splice sites could turn insertions into partial insertions or deletions into partial deletions.
Figure 6. Potential splice junction targets in a typical multi-exon pre-mRNA
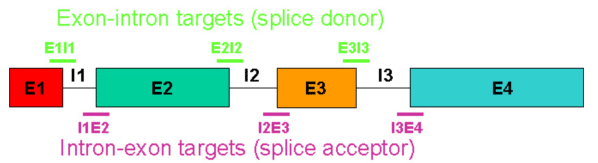
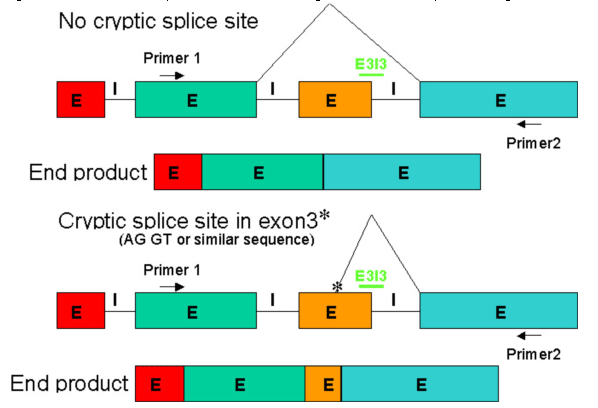
Figure 7 depicts the two most likely outcomes from targeting the exon3-intron3 boundary in the 4-exon transcript. Notice that cryptic splice sites can be activated and may be used instead of splicing to the 5' end of the upstream splice site as predicted in the first example of Figure 7. In general, activation of cryptic splice sites can cause partial deletions or partial insertions.

Figure 7. End products that were observed after blocking E3I3 with a Morpholino oligo
Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis. 2001 Jul;30(3):154-6.

Figure 8 shows a range of possible outcomes of blocking an internal exon, including exon skipping (the most common outcome), intron insertion, and the products of cryptic splice site activation, partial exon deletion and partial intron insertion. The figure shows a Morpholino targeting the donor site; similar outcomes are possible by blocking an acceptor site, though cryptic site activation is less likely because the sequence of a functional acceptor site is more constrained.
Figure 8. Examples of possible products produced by blocking a splice donor with a Morpholino oligo
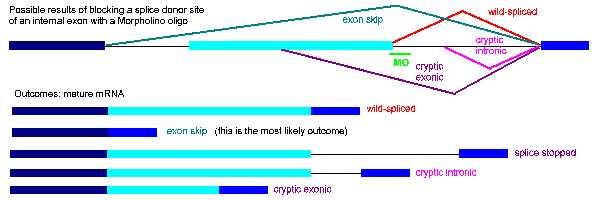

In summary, a Morpholino oligo directed against any splice junction is expected to generate either a complete or partial single exon deletion or a complete or partial single intron insertion. To use RT-PCR to analyze the mRNA product generated by a splice-blocking Morpholino, make a single set of primers targeted to sequence outside (5' and 3') of the predicted deletion or insertion. The Gene Tools customer support staff will be glad to help you both design your oligos and help you determine the best approach for your experiments. Feel free to call or email us.
Limitations: The most significant limitations for using Morpholino oligos fall into two categories, design and delivery. However, the constraints on design and challenge of delivery are only limitations for the unaware.
Regarding design, as long as appropriate regions of the mRNA or pre-mRNA are targeted and problematic moieties such as self-complementarity and runs of G are avoided the oligos are very likely to be effective. But it is important to remember that because Morpholino oligos work by a steric-blocking mechanism, effective oligos must either target sequence to block translation, pre-mRNA modification (e.g. splice jinctions), or regulatory protein binding (e.g. intronic splice suppressors, exonic splice enhancers).
As far as delivery is concerned, the non-charged nature of the Morpholino backbone has historically contributed to difficulty in delivery because Morpholinos cannot be delivered with cationic lipid reagents (lipofection reagents). For cells in culture, delivery of Morpholinos using our Endo-Porter reagent offers good delivery efficacy with less toxicity than when lipofection reagents are used to deliver nucleic acids. Electroporation is also a good method for introducing Morpholinos into cells. Of course microinjection of Morpholinos into embryos has proven to be very effective. It should be noted that delivery has been achieved in nearly all of the most widely studied biological systems. We suggest you look at our database of publications which allows you to browse all organisms studied.
As long as you know the characteristics of good design and the techniques for delivery and plan these into your experiments, we are confident you will achieve the cleanest knockdown available with unsurpassed specificity, minimal non-antisense effects, and amazingly low toxicity.
